
A posteriori dose selection
Source:vignettes/articles/a_posteriori_dosing.Rmd
a_posteriori_dosing.RmdIntroduction
Dosage individualization for a critical care patient treated with amikacin for suspected ventilator-associated pneumonia, using the population pharmacokinetic (ppk) model of Burdet et al. 2015, using the data from therapeutic drug monitoring (TDM).
mod_amikacin_Burdet2015 <- function() {
ini({
THETA_Cl=4.3
THETA_Vc=15.9
THETA_Vp=21.4
THETA_Q=12.1
ETA_Cl + ETA_Vc + ETA_Vp + ETA_Q ~
c(0.1,
0.01 , 0.05 ,
0.01 , 0.02 , 0.2 ,
-0.06 , 0.004, 0.003, 0.08)
add_sd <- 0.2
prop_sd <- 0.1
})
model({
TVCl = THETA_Cl*(CLCREAT4H/82)^0.7
TVVc = THETA_Vc*(TBW/78)^0.9*(PoverF/169)^0.4
TVVp = THETA_Vp
TVQ = THETA_Q
Cl = TVCl*exp(ETA_Cl)
Vc = TVVc*exp(ETA_Vc)
Vp = TVVp*exp(ETA_Vp)
Q = TVQ *exp(ETA_Q)
ke = Cl/Vc
k12 = Q/Vc
k21 = Q/Vp
Cp = centr/Vc
d/dt(centr) = - ke*centr - k12*centr + k21*periph
d/dt(periph) = + k12*centr - k21*periph
Cp ~ add(add_sd) + prop(prop_sd) + combined1()
})
}
mod_amikacin_Burdet2015 <- mod_amikacin_Burdet2015()A posteriori dose selection
Patient record with TDM data
After the first administration, the dosage selection can be refined
using the results of TDM. See
vignette("patient_data_input") for more details regarding
the patient record.
df_patientA <- data.frame(ID=1,TIME=c(0,1,6),
DV=c(NA,58,14),
EVID=c(1,0,0),
AMT=c(2000,0,0),
DUR=c(0.5,NA,NA),
CLCREAT4H=50,TBW=62,PoverF=169)
df_patientA
#> ID TIME DV EVID AMT DUR CLCREAT4H TBW PoverF
#> 1 1 0 NA 1 2000 0.5 50 62 169
#> 2 1 1 58 0 0 NA 50 62 169
#> 3 1 6 14 0 0 NA 50 62 169The concentration measured 30 min after a 30 min infusion do not meet the target for a peak concentration; it is < 60 mg/L.
Estimate the MAP individual parameters
The maximum a posteriori (MAP) individual parameters are estimated.
patA_map <- poso_estim_map(dat=df_patientA,
prior_model=mod_amikacin_Burdet2015)
#> using C compiler: ‘gcc (Ubuntu 13.3.0-6ubuntu2~24.04) 13.3.0’Plot the individual pharmacokinetic profile
The individual pharmacokinetic profile can be plotted using the
rxode2 model provided by the poso_estim_map()
function.
plot(patA_map$model,Cc)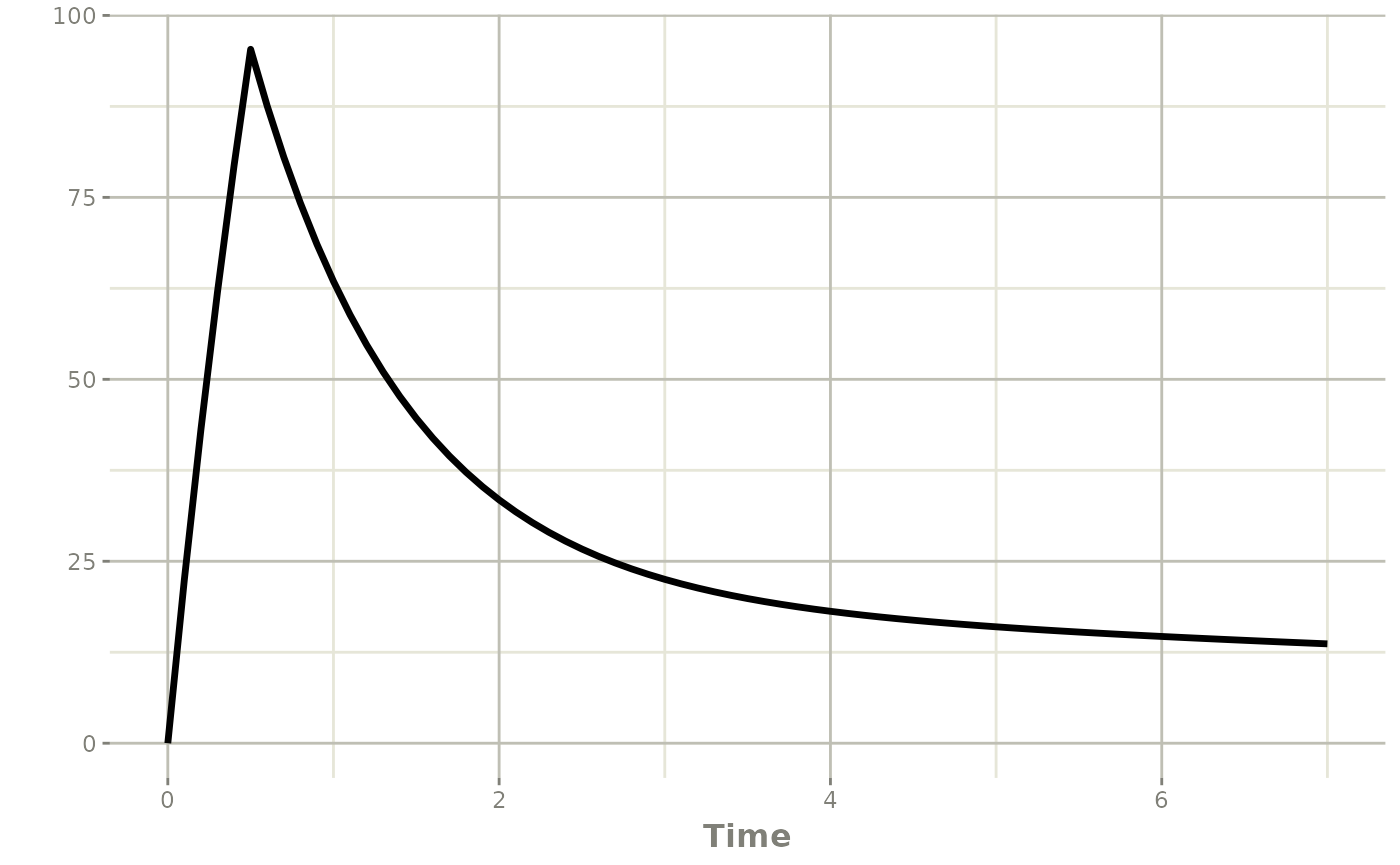
Time required to reach the target Cmin following the first administration
With the MAP estimates of the individual parameters, the prediction of the time needed before reaching the target Cmin can be updated.
poso_time_cmin(dat=df_patientA,
prior_model=mod_amikacin_Burdet2015,
tdm = TRUE,
target_cmin = 2.5)
#> using C compiler: ‘gcc (Ubuntu 13.3.0-6ubuntu2~24.04) 13.3.0’
#> $time
#> [1] 33.9
#>
#> $type_of_estimate
#> [1] "point estimate"
#>
#> $cmin_estimate
#> [1] 2.487844
#>
#> $indiv_param
#> THETA_Cl THETA_Vc THETA_Vp THETA_Q add_sd prop_sd ETA_Cl ETA_Vc
#> 3 4.3 15.9 21.4 12.1 0.2 0.1 0.4499508 0.2730587
#> ETA_Vp ETA_Q CLCREAT4H TBW PoverF
#> 3 0.7061613 -0.1388429 50 62 169The next dose (if needed) can be administered 33.9 hours following the first infusion.
Optimal dose selection a posteriori
The optimal dose to achieve a peak concentration of 80 mg/l can be determined using the MAP estimates.
map_dose <- poso_dose_conc(dat=df_patientA,
prior_model=mod_amikacin_Burdet2015,
tdm=TRUE,
time_c = 35, #target concentration at t = 35 h
time_dose = 34, #dosing at t = 34 h
duration = 0.5,
target_conc = 80)
map_dose
#> $dose
#> [1] 2447.917
#>
#> $type_of_estimate
#> [1] "point estimate"
#>
#> $conc_estimate
#> [1] 80
#>
#> $indiv_param
#> THETA_Cl THETA_Vc THETA_Vp THETA_Q add_sd prop_sd ETA_Cl ETA_Vc
#> 3 4.3 15.9 21.4 12.1 0.2 0.1 0.4499511 0.2730605
#> ETA_Vp ETA_Q CLCREAT4H TBW PoverF
#> 3 0.7061573 -0.1388445 50 62 169The next dose should be 2450 mg.
Interdose interval selection a posteriori
The optimal inter-dose interval to reach a Cmin of 2.5 mg/L before each dosing can be determined using the MAP estimates.
map_interval <- poso_inter_cmin(dat=df_patientA,
prior_model=mod_amikacin_Burdet2015,
dose = map_dose$dose,
duration = 0.5,
target_cmin = 2.5)
map_interval
#> $interval
#> [1] 38.57783
#>
#> $type_of_estimate
#> [1] "point estimate"
#>
#> $conc_estimate
#> [1] 2.500173
#>
#> $indiv_param
#> THETA_Cl THETA_Vc THETA_Vp THETA_Q add_sd prop_sd ETA_Cl ETA_Vc
#> 1 4.3 15.9 21.4 12.1 0.2 0.1 0.4499517 0.2730584
#> ETA_Vp ETA_Q CLCREAT4H TBW PoverF
#> 1 0.7061594 -0.1388427 50 62 169The interval between doses should not be less than 38.6 hours to allow adequate elimination of amikacin between each infusion.